Elhai J, Kato M, Cousins S, Lindblad P, Costa JL (2008). Very small mobile repeated elements in cyanobacterial genomes. Genome Res 18:1484-1499.
|
|
|
|
What kind of repeated sequences are found in a genome? We examined the genome of the cyanobacterium Nostoc punctiforme and tabulated the 20 most numerous repeated sequences. The list included CRISPR repeat units, heptameric tandem repeats, and transposons, but by far the most numerous was a type of repeated sequence that was previously unknown. This class of repeat occurred sporadically in the genome and consisted of units from 21 to 27 nucleotides. We called them Small Dispersed Repeats (SDRs) and identified eight classes of them, SDR1 through SDR8. The sequences of instances of SDR5 are shown in context in Figure 1.
| |
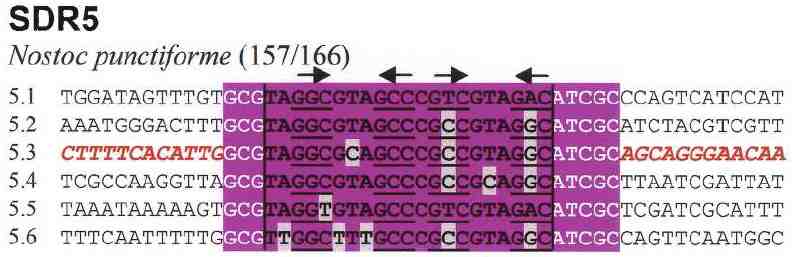
Figure 1. Subtypes of SDR5. The SDR itself is highlighted in purple (deviations from the consensus sequence in gray). The target sequence GCG | ATCGC (HIP1) is shown in white. There are 166 instances of SDR5, 157 of which are in subtypes 1 through 6. |
| | | |
|
Although SDR4, SDR5, and SDR6 differ considerably in sequence, they can each fold into similar structures, shown in Figure 2. The structures are held together by pairing between nucleotides that mutate together. Examples of such compensatory mutations are seen in Figure 1. The structure implies that these SDRs act as single-stranded molecules, presumably RNA and may bind to the same protein.
By comparing regions containing SDRs with similar regions in closely related cyanobacterial strains, it is clear that some of the SDR occurrences represent insertions that occurred in recent evolutionary time. At 21 to 27 nucleotides, SDRs may be the smallest reported mobile elements far smaller than transposons (typically ~1000 nucleotides).
|
|
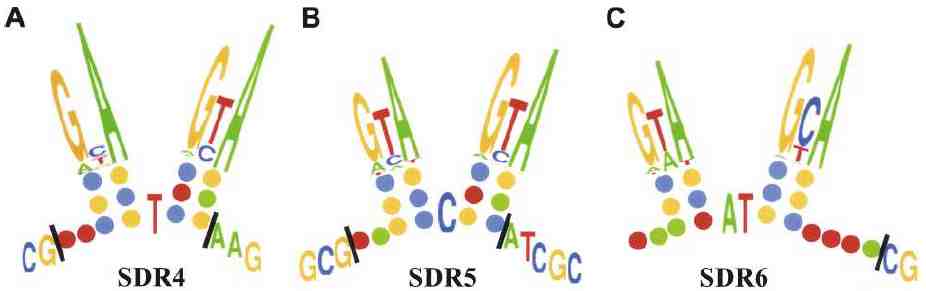
Figure 2. Possible secondary structures for given SDRs. Symbols: green=A, blue=C, yellow=G, red=T. A-T and C-G basepairs are shown. |

