Most think of genomes as collections of genes, but that is only part of the story. In fact, only a little over 1% of the human genome consists of DNA that encodes protein (ENCODE, 2012). The majority of the DNA consists instead of repetitive elements (Richard et al, 2008). To understand our DNA, to understand ourselves, we would clearly be well advised to look into the nature and function of repetitive DNA.
Repetitive sequences can be divided into two broad categories (Figure 1). Tandem repeats consist of DNA units that repeat one after another. The example shown is a portion of the trinucleotide CAG repeat found in the huntingtin gene, which expands to over 40 units in individuals with Huntington's disease (Cattaneo et al, 2002).In contrast, the units of dispersed repeats are situated distant from one another. Transposons are well known examples of this class, but there are instances that are much smaller -- sequences of just several nucleotides in length that may appear far more frequently than expected by chance.
It would be useful to address the following questions:
- What are they? What are the sequences of the repeating unit? How frequent are they? Where do they occur?
- Where do they come from? Are they copied from an existing source of repeating units? Do they arise by mutation? Are they conserved among related organisms?
- What do they do? Are their clues that indicate a function that aids the cell? Are they more likely to be purely selfish entities, propagating themselves without conferring advantage on the host? Something in between?
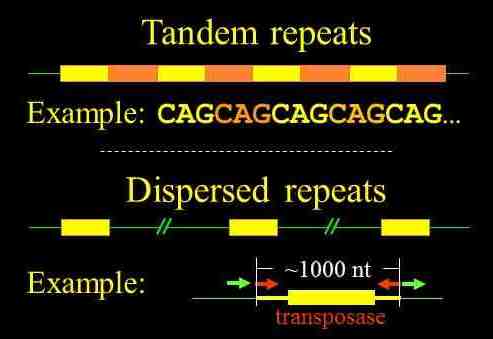
Figure 1. See text for explanation. The gene encoding the transposase (which is responsible for the movement of the transposon) is flanked by inverted repeats (red arrows) that mark the termini of the transposon and direct repeats (green arrows) generated by transposition.