
Second, it can count the number of times a string (a group of characters) appear in another string (including a DNA sequence):

If every ATG marks the beginning of some gene (though all genes do not necessarily begin with ATG), then the number of ATG's should be no more than the number of genes. Is it?
Let's see if there is a similar indicator preceding genes, but for some variety, switch to the freshwater cyanobacterium Synechococcus PCC 7942 (abbreviation S7942B). First verify that genes of this organism start with the same triplets as the genes of ss120.
You might be able to find some sort of pattern in all those nucleotides (the human mind can find a pattern in anything), but certainly nothing jumps out as did the initial triplets.
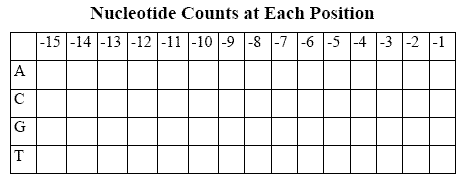
Some statistical analysis might focus our attention on areas of interest. Suppose we built a table that looked like what you see to the right.
If there were particular biases for or against certain nucleotides at specific positions before genes, maybe the table would make them apparent. Lets find out. First, give the set of sequences a name, something like:

Not something you want to do when the weather's nice outside. Fortunately, BioBIKE can do this automatically, using a function called MAKE-PSSM-FROM. A PSSM (Position-Specific Scoring Matrix) is a table of the sort we imagined, except that frequencies instead of counts are given.
Find this function in the STRING-SEQUENCES menu and Bioinformatic-tools submenu, giving you in the end something like the following:

You could click on the aligned-list gray argument box and type in the name upstream-sequences, but here's a more foolproof method: click on the gray box, then click on the VARIABLES menu, and finally click on upstream-sequences. That will transfer the name of the variable to the argument box with no possibility of misspelling error. Finally, execute the function. You will get a message indicating that you generated a two-dimensional table, as expected.
