Welcome to BioBIKE's tutorial
What is a Gene?
What is the beginning of a gene?
Maybe you can find the beginning of a gene in a chromosomal sequence, but cells don't have the advantage of a nice coordinate list. How do they do it? How do you find the beginning of a gene -- a bunch of nucleotides -- in the middle of a genome -- an even bigger bunch of nucleotides?
No idea? Let's start over. Maybe you can find the beginning of a sentence in a book,... How do you do it? How do you find the beginning of a sentence -- a bunch of symbols -- in the middle of a book -- an even bigger bunch of symbols?
That's not so difficult... how do you figure out where sentences begin? If you examine your internal processes, perhaps you'll come up with two types of strategies:
- Look for an internal cue -- i.e. a capital letter. Words with capital letters are candidates to begin sentences.
- Look for an external cue -- i.e. punctuation. Words following periods or question marks are candidates to begin sentences.
Perhaps genes have internal or external cues. Let's look....
... but first, you might want to clear away some of the debris on your screen. You can click the red X box at the upper left of each function you want to get rid of. I suggest keeping three: SEQUENCE-OF pro0029, COUNT-OF..., and DESCRIPTION-OF.... They may come in handy later.
|
- You can stare all you want at a single gene, but unless you have prior knowledge (like what the equivalent of a capital letter looks like in DNA sequences), it won't do any good. What we need to do is to look at many genes and see if we find any general features.
So let's grab the first, say, 10 nucleotides of every gene ss120 has and examine the collection to see if anything pops out. First, you'll see how to do this with one gene, then you'll generalize to all. Try this:

You can extend the DISPLAY-SEQUENCE-OF pro0029 box you may already have on the screen. If it's not there, then you can get the function from the GENES-PROTEINS menu. Add the From and To options from the option menu. Don't fall prey to a very common error of typing in numbers and forgetting to click Enter!
Are these in fact the first 10 nucleotides of pro0029?
Computers are a source of power, but they're also sly and evil. They will fool you every time they can, and you must constantly be on your guard. Any time you have a chance to check their work by hand, DO IT.
|
From your the work you did in the preliminary section, check at least for consistency, a confidence-building measure that the function is telling the truth.
- OK, so it's telling the truth (this time). If the function can work with one gene, it can work with all genes. Modify the function SEQUENCE-OF function, replacing pro0029 with all genes of ss120. Here's how. First, erase the previous argument, pro0029, by clicking the magenta X at the upper right corner of the argment box:

This will clear the argument box.
Now click on the argument box to select it and bring in GENES-OF from the GENOME menu. Type ss120 in the argument box of GENES-OF (click Enter), getting:
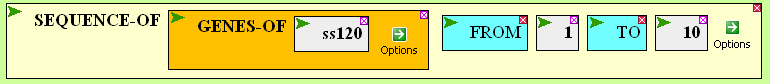
Now execute the command again. To see the output in its full glory, click on the screen icon in the upper right corner of the results window:
Actually, click it again to fully expand the window (You'll click it again in a moment to toggle it back to small size).
- Notice the three dots at the end... BioBike has again saved us from generating an overwhelming amount of output. It puts only the first 100 elements of a list on the screen. Note the "..." at the end of the list. (You can modify this number by changing the preferences accessible through the FILE button) (but don't -- 100 is enough). What fraction of the gene beginnings were listed? (If you don't know the answer, what fact would you need to know to figure out the answer?)
- Consider the results and -- something only humans can do -- find something interesting, a pattern of undefined nature you think is important.
No doubt you see something, a general tendency that holds for only the first three nucleotides. Let's focus on them. Toggle the results window back to a small size through the window icon and revise the SEQUENCE-OF command and execute it, to generate a list of the first three nucleotides of each gene in ss120.
- Time to come up with a hypothesis. What single three-nucleotide sequence looks like it might serve as the capital letter (or perhaps a capital letter) that marks the beginning of sentences? Of course, a hypothesis that isn't tested is pretty useless. What could you do to test your hypothesis, given just the resources at your disposal?
You're looking at the beginnings of only the first 100 genes, a small fraction of the whole. Maybe they're not representative? Can you check this?
To do so, it would be useful to determine the fraction of genes that begin "ATG". You need to be able to COUNT the number of genes beginning "ATG" and divide that by the total number of genes. You can do this with a variant of a tool you've already run across. Go to the LIST-TABLES menu, then List Analysis, and get COUNT-OF. Type "ATG" (be sure to include the quotes) into the gray argument box...
...but that's not enough. You're looking at the list of three nucleotides, but you need to tell BioBIKE that this is the list you want to count. Mouse over the options arrow and click on the option IN.
Now you're in what should be familiar territory. You could copy or cut the command that produced the list and paste it into the gray object box governed by IN. You could... but that would be ugly and inefficient. Why ask the computer to calculate something that it has already calculated?
A better idea is to give the list a name. To do this, go to the DEFINITION and click on DEFINE. Now click on the variable-name box and type in the name you want to give the list. It could be almost any name you like (so long as the name doesn't have an embedded space). Press Enter. Click the value box and... how to refer to the list you generated? There are a few ways. One way is to go to the OTHER COMMANDS menu and select PREVIOUS-RESULT. You should end up with something like:

Execute this. Note that a new button, VARIABLES, now appears in the pallette.
- Return to the still incomplete COUNT-OF command, click on IN's gray object box to select it. Now you can go to the VARIABLES menu and click on the variable you just made, bringing it down to the object box. If you execute the resulting function, you should learn how many genes in ss120 begin with "ATG".
- How do you interpret the answer? How does the number compare to the total number of genes?
- The list of triplets isn't all "ATG". Modify the COUNT-OF command to repeat this with other triplets you think may be initiating elements.
- Add up all the counts and compare them to the total number of genes. Conclusion?
- Evidently, there are many exceptions to the rule. The greatest insights are often gained by investigating exceptions. Can you identify any gene of ss120 of the first 100 displayed that is one of the exceptions? Presume that the list of triplets goes in the same order as the list of genes. Once you have identified the gene, use DESCRIPTION-OF in the GENES-PROTEINS menu to find out more about it (use the FULL option).
- Take a look at the Annotation field of the gene: It says the gene encodes an RNA! What could that mean? Don't all genes make RNA, which is then translated into protein?
(Lost? If so, check the name of the gene whose information you were given. If it is A7120.ffs... TREACHERY! It should be PRO1375.ffs, not A7120.ffs. As it happens two organisms have genes of the same name, and the computer, predictably, chose the one that would fool you. Repeat the command using the full name, pro1375.ffs)
|
Another thing, the field Encodes-protein does not have the value of True! (NIL is BioBike speak for false). Are there genes that do not encode protein? It's time to call in reinforcements.
Go to Google and type in "signal recognition particle RNA". The first site listed should be SRPDB Welcome. Go there and take a look at the overview
(click on About SRP). Learn something about the SRP cycle – what is it? What is SRP made of?
- OK, back to business. Evidently the ffs gene does not encode a protein, and it does not begin with one of the usual triplets. Is that true of other genes that don't encode proteins… wait a second. Are there other genes that don't encode proteins, and if so, how many? We need to find out more about ss120. Modify the DESCRIPTION-OF to get a description of ss120.
- Scrolling down you'll see a (partial) list of genes in ss120, and further down, sure enough, there's a list of noncoding genes. How many are there? Time for a new hypothesis concerning the beginning of genes. What can you come up with?
- Test the hypothesis: What triplet(s) do the noncoding genes begin with? You got the first three nucleotides of all genes of ss120. How can you modify that command to confine it to the noncoding genes?
Typical scenario:
- Define a problem
- Imagine a tool that would solve it
- Look for such a tool... maybe someone else has encountered the same problem
- If not, build the tool
|
Look through the tool box in the GENOME menu and see if something doesn't commend itself.
PROBLEM 2:
Find the first three nucleotides of every gene in ss120 that does not encode a protein. Propose a generality that explains as much as possible of what you've found thus far. What does it explain? What doesn't it explain?
PROBLEM 3:
You probably saw some common sequences while answering PROBLEM 2. Is there any generality you might draw concerning the beginnings of noncoding genes? Recall that you discovered noncoding genes before by finding a characteristic shared by genes that don't have the usual start sequences. Are there other characteristics within noncoding genes that might help you make further connections? Use DESCRIPTION-OF to explore genes you think may be connected.
A good start!
Use your browser to go back
one page to the table of contents.




